Microbial Response to Crude Oil in Ocean Sediments Using ‘Omic Technologies'
Connect with an expert opens in a new pageChallenge
The Solution
The Outcome
- Systems biology approach incorporating multidisciplinary assays
- Extensive evaluation of microbial communities with depth previously unattainable
- Rapid screening of complex environmental samples
- Unbiased detection relative to culturing in laboratory
- Important gene and biochemical pathway identification
- Key protein identification/quantification
- Discovery tool, capable of detecting the unexpected
Integrating next-generation sequencing (NGS) data with functional protein profiling approaches allows for comprehensive identification of diverse and difficult to culture microbes in environmental samples. The types and frequencies of detection of hydrocarbon-degrading bacteria identified by Battelle scientists using this approach are tabulated below.
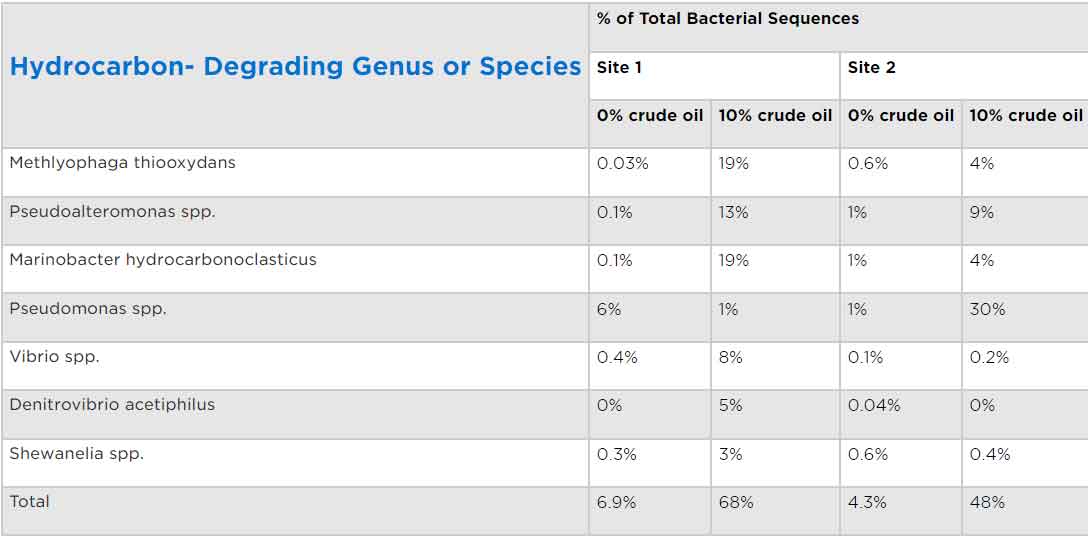
Table 1. Frequency of detection of bacterial species in different environmental samples with and without exposure to crude oil. Microbial metagenomics analyses were conducted on sediment samples from two geographically distinct nearshore sites. The samples were then incubated with 10% crude oil and the frequencies of bacterial sequence detection compared. A ten-fold increase in detection of genomic sequences from bacterial species with known hydrocarbon-degrading capabilities was observed in the samples from both locations when the samples were incubated with 10% crude oil [6.9% (no-oil control) to 68% (with oil) in the Site 1 sediment, and 4.3% (no-oil control) to 48% (with oil) in the Site 2 sample]. Interestingly, while the same hydrocarbon degrading species were initially present in both sediment samples, incubation with 10% crude oil led to increased abundance of Methylophaga thiooxidans, Pseudoalteromonas spp., and Marinobacter hydrocarbonoclasticus which were dominant in Sample 2, while Pseudomonas spp. became most abundant in Sample 2. The test illustrates how indigenous microbial communities respond to crude oil exposure in sediments from different geographic locations.
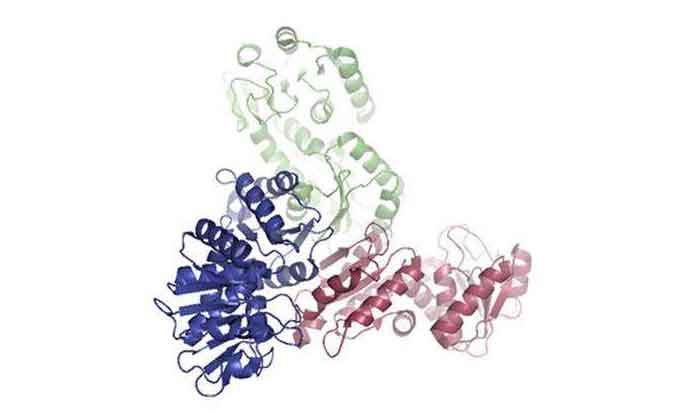
Figure 1. A 3D image of the protein methylene-tetrahydrofolate dehydrogenase, an oxioreductase, which was only identified from oi-treatment sediments. This enzyme is involved in the degradation of organic materials.
Case Studies
Technology Commercialization and Licensing
With over 90 years of research and development experience, Battelle supports all areas of industry with intellectual property licensing focused on bringing our cross-disciplinary expertise to meet our clients' most difficult challenges.
